DNA Structure
1. Nucleotides
Deoxyribonucleic acid, or DNA, like proteins, is a linear macromolecule found in all living cells. In contrast to proteins, however, it is build up of only 4 different types of building blocks, called nucleotides. Nucleotides are composed of a base, being either a purine or pyrimidine group, and a 2'-deoxyribosyl-tri-phosphate. The four types of bases composing the sequence of DNA are:
Purines:
Adenine A 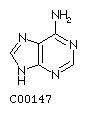 Guanine G
Guanine G 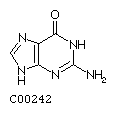
Pyrimidines:
Thymine T 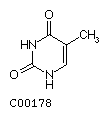 Cytosine C
Cytosine C 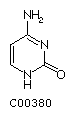
(Note: more structures can be found at KEGG database)
The sugar is a 2'-deoxy ribose which is phosphorylated at its 5' hydroxyl group. Free nucleotides contain either one, two or three phosphates representing the mono-, di-, or triphosphate form of the nucleotide, the latter being known as dATP, dGTP, dTTP, and dCTP.
Fig. dATP
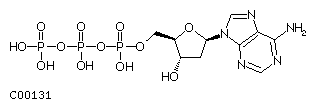
Nucleotides are covalently linked in DNA through their 5'-phosphate to
the 3'-hydroxyl of the next nucleotide. The linear polymer is synthesized
in the 5' to 3' direction (the 3’-OH nucleophilically attacks the Pa
of the incoming nucleotide) and the deoxyribose phosphate backbone is
a polyanion at physiological pH, i.e., it carries multiple negative
charges because the pKa value of the phosphodiester bond is
close to 1.
2. B-DNA or Watson and Crick double helix
Single DNA strands are not stable, but associate with a second strand
to form a double helix structure, where both strands intertwine around
each other. At the center of the helix the four bases are H-bonded to
each other and they do so in a very specific way. The bases point toward
the helix center and H-bond in the following manner only:
G with C 3 H-bondsA with T 2 H-bonds
The rigidity and linear geometry of the H-bonds restricts base pair (bp)
formation. The plane of the base pair lies perpendicular to the helix
axis. The right handed B-DNA form is the physiological form of the DNA
double helix as first described by Watson and Crick in 1953. The specific
conformational constraints made it clear that the sequence of the bases
in the polymer encodes the genetic information for the synthesis of proteins.
The B-DNA is a right handed, anti-parallel double helix with 2nm
in diameter.
10bp per turn
a twist of 36° per bp
0.34nm (3.4Å) per base
pitch 3.4nm (34Å)
diameter 2.0nm
base plane tilt away from being perpendicular to helix axis 6°
The B-DNA helix as described by the above parameters represents an idealized
helix. The conformation of 'real' DNA, however, deviates slightly from
the B form in a sequence dependent manner as well as depending on the
interaction with DNA-binding proteins. Examples of protein - DNA interaction
mechanisms will be discussed in sections 3.1 and 3.2. An important feature
of the double helix are the minor and major grooves,
winding along the helix surface. In those grooves, parts of the aromatic
ring structures of the purines and pyrimidines are exposed to the surface
of the helix. The major groove is the site where most protein-DNA interactions
occur.
Fig.. B-DNA sideview
and projection along helical axis
Incidentally, it was the structure of the a -helix in proteins that led most structural biologist to look for an analogous structure in nucleic acids, i.e., helical or double helical structures with the bases sticking away from the helix center as do the residues in protein helices. It was the insight to model the base pairing inside the double helix that was truly revolutionary about discovery of the DNA structure. (See the reprints of the two original papers by Watson and Crick at the end of this section.)
A measurable quantity of DNA is its contents of G+C and A+T base pairs (besides the sequence per se) which differs from organisms to organism (Chargaff rule) and is characteristic of an organism's genome. The G+C content in mammals varies from 39 to 46% and in bacteria from 25 to 75%. The G+C content exhibits an important correlation with the stability of the double helical conformation of DNA due to the difference in hydrogen bonding capacity of GC pairs with 3 H-bonds and AT pairs with 2 H-bonds. This has been shown through heat denaturation of the double helix into a single strand conformation. So called melting curves can be measured by increasing the temperature of the solution. At a certain temperature, called melting temperature or Tm, 50% of the DNA is found in double strand (ds) and 50% in single strand (ss) form. The melting temperature is directly proportional to the GC content.
Fig. Denaturation behavior of DNA

A conformational change can be induced in DNA when the relative humidity
of the sample is lowered below 75%. The double helix forms now what has
been described as A-form. The A form is a wider and flatter helix, the
base pair plane is tilted with respect to the helix axis, the helix also
exhibits a minor and major groove with the following structural parameters:
11bp per turn
a twist of 33° per turn
0.26nm per base
pitch of 2.8nm
diameter 2.6nm (has an axial hole)
base plane tilt away from being perpendicular to helix axis 20°
Fig. A-DNA dimer
(acutally an RNA duplex with parameters identical to A-DNA)
5. Z-DNA
This helix has a left handed conformation with the following structural
parameters:
12bp per turn
0.37nm per base
pitch 4.5nm
diameter 1.8nm
base tilt 7°
The biological significance for both A and Z helices is not well understood.
Structural studies of Z-DNA are derived from synthetic oligonucleotides
rather than long stretches of DNA extracted from cells.
Fig. Z-Dna Hexamer With 5' Overhangs That Form A Reverse Watson-Crick
Base Pair
projection along helical axis:
Note: To view DNA structures above from the Brookhaven
Protein Database search for keywords or use the followoing accession numbers:
355d or 311d for B-DNA; 104d for a DNA/RNA duplex; 309d for A-DNA, and
312d for Z-DNA.
6. Genomic size of DNA
Consider the size of DNA and its structure of being a long, linear polymer. In prokaryotes, the entire genome consists of one single molecule, mostly in circular form. As mentioned in the Introduction, the total genome of Haemophilus influenzae has been sequenced containing more than 1.8 million base pairs. Often DNA is found to be methylated, the result of an enzymatic modification that serves as chemical protection. Methylated bases can often not be recognized by DNA binding proteins such as nucleases that hydrolyze the phosphate ester bond between nucleotides. In this way, bacteria protect their own DNA from degradation, whereas the DNA of intruders (like viruses) which is not methylated can be degraded. Methylation occurs at:
N6-methyl-dA
5-methyl-dC
Because of the large size of circular DNA, the helix is often wound up
in a super-coiled form. The combination of supercoiling and helix
conformation has an effect on the recognition of DNA binding proteins
(it changes the local conformation of the B-DNA double helix).
1. Ribonucleic acid
RNA is very similar to DNA in that is made of 4 different building blocks, the ribonucleotides. The pyrimidine base thymine is modified in that it lacks a methyl group and the resulting uracil takes its place in base pairing. The ribose comes in its fully hydroxylated form. Together, the presence of uracil in place of thymine, and the 2'-OH in the ribose constitute the two chemical differences between RNA and DNA. RNA is composed of the four bases:
Purines:
Adenine A  Guanine G
Guanine G 
Pyrimidines:
Uracil U 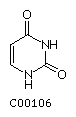 Cytosine C
Cytosine C 
(Note: more structures can be found at KEGG
database)
2. RNA structure
RNA differs, however, from DNA because it does not form an analogous
double helical structure. RNA does, however, form base pairs with DNA
resulting in a heteromeric double helix consisting of one DNA and one
RNA strand. This annealing of an RNA strand to its complementary DNA strand
is called hybridization and plays a crucial role in the transcription
and translation of genetic sequences into protein sequences. RNA does,
in contrast to DNA, form short double strand structures on itself, thereby
forming so called stem and loop structures. Both DNA/RNA double helices
and RNA/RNA double strands have an A-DNA like conformation, also called
A-RNA or RNA-II. The helix has
11bp per turn
pitch of 3.0nm (30Å)
There are three major RNA species that can be distinguished both on their
ability to form stem and loop structures as well as their functional role
in the cell. The three types of RNA are:
messenger RNA or mRNA
transfer RNA or tRNA
ribosomal RNA or rRNA
In addition there are RNA molecules found in viruses (viral RNA) that
serve as the genomic blue print that normally is encoded in DNA, and ribonucleo-proteins
of diverse origin both ribosomal and non-ribosomal in nature.
Before discussing the individual types of RNA, let's have a look at the cellular machinery involved in protein synthesis. This diagrammatic overview is helpful in understanding the distinction of those RNA types. In both prokaryotes and eukaryotes, protein synthesis is very similar in its hierarchy, but differently organized in spatial arrangements and intra-cellular location. Prokaryotes, having no nucleus, transcribe the DNA sequence into mRNA, and the mRNA sequence is translated into the protein sequence with the intermediate tRNA molecules, which have the anti-codon information covalently linked to the corresponding amino acid. The ribosome is the place where mRNA and aa-tRNA come together and protein elongation takes place. As the diagram shows these can be simultaneous processes, i.e., the emerging mRNA strand is immediately translated into the polypeptide.
In eukaryotes, transcription, or synthesis of mRNA, and translation,
the synthesis of proteins, are separated in space and time. Transcription
occurs in the nucleus, where the mRNA is synthesized and processed (splicing).
The resulting mRNA is transported to the cytoplasm and translated into
a polypeptide, in an identical process as found in prokaryotes.
3. Transfer or tRNA
This section will only deal with the structure of the smallest of all RNA species, the transfer RNA. The tRNA molecules are key to the translation process of the mRNA sequence into the amino acid sequence of proteins (at least one type of tRNA for every amino acid). To be precise, the amino-acyl-tRNA-synthase proteins are the 'true' translators of the genetic code into an amino acid sequence. These synthetases acetylate tRNA molecules with the proper amino acid that corresponds to the anti-codon in the structure of the tRNA molecule. The anti-codon later recognizes the codon, the triple base sequence which 'codes' for the amino acid along the mRNA strand. A failure of properly acetylating the tRNA with the right amino acid results in a amino acid mutation even though the DNA sequence has not been changed.
tRNA molecules are small nucleic acids of 60-95 nucleotides, mostly 76,
with a molecular weight 18-20kD, with the secondary structure resembling
a clover leaf. Here are a few common features shared by all tRNA molecules
found in various organisms.
(1) 5' terminus always phosphorylated
(2) 7 bp stem, may have non-Watson&Crick pairing (like GU)
acceptor or amino acid stem at 3' terminus, last three nucleotides CCA-3'-OH,
amino acylation occurs at 3'-OH group of
(3) 3-4 bp stem and loop contains the base dihydrouridine (D) [D- arm]
(4) 5 bp stem and loop containing anti-codon triplet [anti-codon arm]
(5) 5 bp stem and loop contains sequence TY C, Y standing for [T- arm]
pseudouridine
(6) variable arm (between anti-codon and T arm) of length 3 -21 nucleotides
(7) contains numerous modified bases (up to 25%) which are all post-transcriptionally modified
The three dimensional structure of tRNA resembles an L-shaped molecule
with the D-arm and anti-codon loop building one stretch and the T-arm
and acceptor stem building the other stretch being deposed by ~90°
to one another (interstem angle of 82°
by X-ray refinement and 92° in an electron
microscopy study[Hagerman and Amiri, 1996]). The molecule is about 6 nm
in each direction with the anti-codon to acceptor 3'-term ends being 7.6
nm apart. The diameter of both arms are about 2.0 to 2.5 nm.
Fig. Rasmol structure of tRNA from yeast
Note:Acceptor stem loop upper left; anticodon loop bottom; To view RNA structures from the Brookhaven Protein Database search with keywords or use accession number 1SLO (first stem loop of S11 ribosomal RNA of C.elegans, NMR structure)
The structural complexity of tRNA is reminiscent of that of a protein
with 71 out of 76 bases participating in stacking interaction (of which
42 in double helical stem structures). 9 bp interaction are cross linking
the tertiary structure, i.e., they interact with bases from a different
stem and loop region. All of these 9 bp are non-Watson-Crick associations
and are highly conserved which makes it likely to predict similar structures
for all tRNA molecules (in fact, only few tRNA molecules have been crystallized
and their structure determined).
4. Codon - anti-codon interaction
The genetic code is made such that always three bases in a row, a triplet, codes for a specific amino acid. Thus a sequence of triplets in the DNA can be transcribed into a sequence of triplets in the mRNA strand. Since the DNA is a double helix formed by two complementary strands, the anti-sense strand is transcribed into mRNA resulting in the +sense on the mRNA level (with U instead of T).
As a rule, every tRNA can be covalently linked to only one type of amino acid, through the specificity of the amino acyl tRNA synthase. A specific amino acid can be linked to different tRNAs. These tRNA molecules are called iso-accepting tRNAs. There are as many tRNA species as codons used for translation.
Many tRNAs bind to two or three of the codons specifying their amino acid. This happens through non-Watson-Crick base pairing at the third codon-anti-codon position and is responsible for the degeneracy of the genetic code. This not-so-precise base pairing is referred to as the wobble hypothesis and is due to the presence of a modified base in the anti-codon structure, namely a methylated guanosine, Gm, or I. For example the tRNA for phenylalanine (PhetRNA) binds two codons in the mRNA:
tRNA anti codon
3' A A Gm5' 3' A A Gm5'
½ ½ ½
½ ½½
mRNA codon
5' U U C 3'
5' U U U 3'
The Standard Code (follow the link to the genetic code tables at NCBI)
By default all transl_table in GenBank flatfiles are equal to id 1, and
this is not shown. When transl_table is
not equal to id 1, it is shown as a qualifier on the CDS feature.
TTT F Phe TCT S Ser
TAT Y Tyr TGT C
Cys
TTC F Phe TCC S Ser
TAC Y Tyr TGC C Cys
TTA L Leu TCA S Ser
TAA * Ter TGA *
Ter
TTG L Leu i TCG S Ser
TAG * Ter TGG W
Trp
CTT L Leu CCT P Pro
CAT H His CGT R Arg
CTC L Leu CCC P Pro
CAC H His CGC R Arg
CTA L Leu CCA P Pro
CAA Q Gln CGA R Arg
CTG L Leu i CCG P Pro
CAG Q Gln CGG R Arg
ATT I Ile ACT
T Thr AAT N Asn
AGT S Ser
ATC I Ile ACC
T Thr AAC N Asn
AGC S Ser
ATA I Ile ACA
T Thr AAA K Lys
AGA R Arg
ATG M Met i ACG T Thr
AAG K Lys AGG R Arg
GTT V Val GCT A Ala
GAT D Asp GGT G Gly
GTC V Val GCC A Ala
GAC D Asp GGC G Gly
GTA V Val GCA A Ala
GAA E Glu GGA G Gly
GTG V Val GCG A Ala
GAG E Glu GGG G Gly