Self - Assembly Systems
1. Introduction
Self-assembly systems are defined by the regular assembly of small molecular entities into larger supra-molecular structures exhibiting new functions that can not be exhibited by the isolated monomer. The interaction between subunits are due to non-covalent, electrostatic forces. For cellular systems the driving force for assembly is the hydrophobic effect. At the most basic level all living organisms are self-assembling entities. The following is a selection of biological systems to illustrate the role of self-assembly mechanisms for the sustenance of life.
The balance between functional formation of supra-molecular structures
(membranes, cytoskeleton) and those which are destructive (S hemoglobin
in sickle cell anemia) are often based on minute changes in surface composition
of the monomer. Replacing polar with non-polar, or non-polar with charged
residues, affects the electrostatics of the interaction between subunits.
This is nicely demonstrated by the effect of a single amino acid mutation
in S-hemoglobin (sickle cell anemia). Phospholipids which form stable
planar membrane structures can be disrupted locally by the self-assembly
like insertion of antibiotic peptides such as gramicidin and mellittin,
to mention two.
2. Phospholipids and Detergents
In aqueous solution, phospholipids aggregate to form larger micellar and membranous structures due to their solubility properties in water. Cell membranes are the most common found supra-molecular structure formed by phospholipids, which form 2-dimensional matrices for the assembly of membrane proteins and lipids such as cholesterol, which by themselves would not form planar membrane structures.
The combination of hydrophobic and hydrophilic groups in the same molecule
is a common theme in phosopholipids and detergents and an important structural
feature determining their assembly into either membranes or micelles,
respectively. The ratio of the size between head group and 'tail' determines
the solubility in aqueous solution and its aggregation behavior (hydrophobic
effect). The buried hydrocarbons interact with each other through Van
der Waals forces, while the polar headgroups are hydrated in water. The
aggregation number of self-assembled monomers within micellar aggregates
is characteristic for each type of amphipathic molecule and ranges from
10 to 100 molecules. The experimental value associated with these aggregates
is the critical micellar concentration (cmc). The cmc value gives
an estimate of the concentration of free, monomeric molecules in solution
above which they start to aggregate. The cmc defines the saturation
solubility of amphipathic molecules.
Phospholipids are amphipathic molecules. Put into water, they form micelles and predominantly spherical membrane structures called liposomes or vesicles. At the air-water interface phospholipids form single layers, or monolayers, with their polar headgroups solvated in water and their hydrocarbon tails sticking into the air (non-polar or low dielectric medium).
Fig. Monolayer of phospholipids at air/water interface
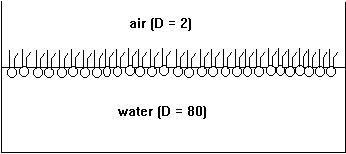
The formation of micellar and vesicular structures (and monolayers, too) is the consequence of the hydrophobic effect. The large hydrocarbon tails aggregate in the center of the particle excluding water from contact with the non-polar structures (dielectric constant D=2). The charged, polar headgroups form the particle's surface making it perfectly soluble in water (dielectric constant D=80). Membranes can be formed in the laboratory simply by mixing lipids with water (normally a buffer or electrolyte solution, containing salts and a buffer substance to stabilize the pH of the solution). Techniques such as ultra-sound sonication and repeated freezing and thawing of phospholipid solutions produces vesicles that contain one or several layers of membranes, also called phospholipid bilayers because they are formed from two opposing monolayers.
Fig. Schematic diagram of a membrane, or phospholipid bilayer, with negative surface charges (-).
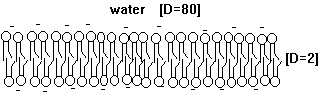
In the plane of the membrane lipids are free to move, they can diffuse laterally. This diffusion is quite fast and in 1972 biological membranes have been described by the fluid mosaic model. Even though lateral diffusion is appreciably high, the flipping of phospholipid units from one monolayer to the opposite layer is a very rare process and large energy barriers are the major reason that restrict the movement of polar headgroups across the non-polar core of the bilayer. In cell membranes, lipids are transported from one monolayer to the other by enzymes, so called flippases, a process which is powered by ATP hydrolysis.
Biological membranes are not only composed of lipids, but also contain proteins, or membrane proteins. This mixture of different types of macromolecules in a stable, flat structure, was recognized as being rigid as well as flexible, and the components were distributed unequally, dependent on their lateral mobility. Although in a cell, many proteins are responsible for the stability of the membrane and the distribution of its components. It is the self-assembly properties of lipids and membrane proteins in an aqueous environment that causes membrane formation. The driving force of this self-assembly process is the hydrophobic effect, i.e., the fact that non-polar parts of a molecule aggregate with each other to exclude water molecules. The hydrophobic effect is mostly an entropic effect because the water molecules that would surround the hydrocarbons could do so only by decreasing the entropy of the water shell structure (more ordered). The self aggregation of hydrocarbons frees water molecules and gives the water structure a higher entropy, which, according to the second law of thermodynamics, indicates the direction of a spontaneous process.
Like phospholipids, membrane proteins are amphipathic structures. The
hydrophobic part of the protein is embedded in the hydrocarbon part of
the lipid membrane, and hydrophilic surfaces of the protein are in contact
with the lipid headgroups and the water surrounding the two sides of the
membrane. This arrangement gives membrane proteins a particular property,
namely their orientation is important for their function. Membrane proteins
can control vectorial processes.
4. Channel forming peptides
Gramicidin A - a bacterial antibiotic peptide
Gramicidin A is a small, 15 amino acid containing peptide that incorporates into cell membranes and forms a dimer in a head-to-head interaction of two individual helices. This dimer formation is spontaneous and stabilized by H-bond formation and constitutes the simplest possible self-assembly system. The peptide is both structurally and functionally well understood. The kinetics of dimer formation and dissociation has been studied extensively. The monomeric form is inactive and the dimer is the active form (forming an ion channel across cell membranes thereby depleting the ion gradient across these membranes leading to cell death).
Mellittin - bee venom peptide
Mellittin is a 23 amino acid containing peptide that incorporates into cell membranes by aggregating into a -helical bundles, called alpha barrels, of different sizes. Each peptide is long enough to span a phospholipid bilayer and the helical bundle forms a central pore where ions can pass across the membrane. This causes a depletion of the ion gradientwith the size of the pore not being defined as in Gramicidin A.
The free energy terms (D G in kcal/mol) of separating two peptides (AB ion pair) that interact through a salt bridge in either aqueous or non-polar environment are shown in the figure below [Engelman, D.M., 1982]. A dimer of two a -helical peptides, one carrying a single positive and the other a single negative charge in the center of the helix, is not stabilized in water because of the hydration of the ionized groups of groupa A and B. The ion pair is stabilized in a hydrophobic environment like a membrane (see section 1.1 for details). Hydrogen bonds contribute also to the stability of the heterodimer in membranes, whereas the conformational entropy term of the peptide is equal in both cases.
The figure also shows D G values for dimers interacting through hydrophobic groups and strong or weak hydrogen bonds. As can be seen by the D G = -5kcal/mol of the conformational entropy term upon separating the dimer into monomers, the dimer restricts restricts rotational degrees of freedom as compared to the monomeric form.
Fig. Free energy effects in the separation of twenty residue a
-helices in aqueous and non-polar environment
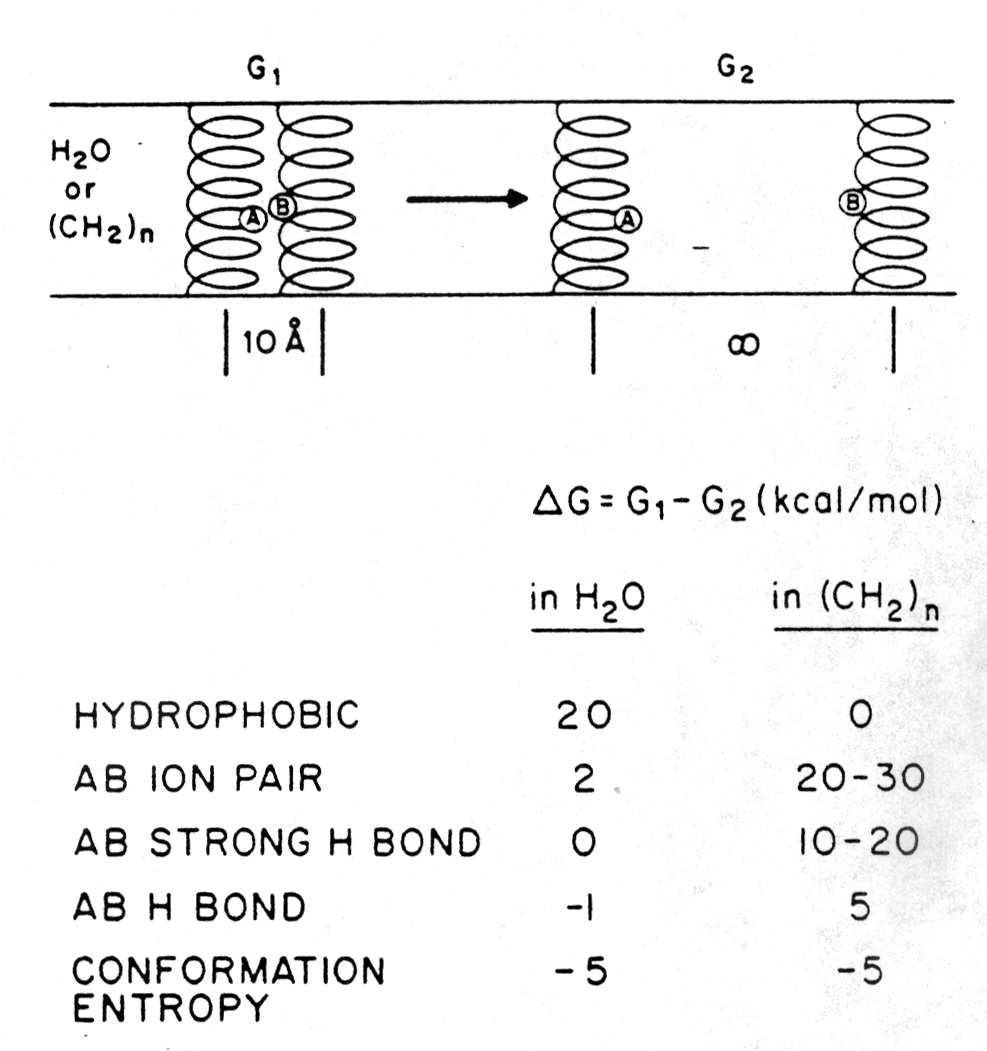
from Engelman, 1982
5. Protein Filaments: S-hemoglobin - Sickle Cell Anemia
Hemoglobin is normally found in a tetrameric state that is necessary
and sufficient for oxygen binding. A specific single point mutation that
replaces Glu6 for Val6 in the b gene induces
the aggregation of mutant hemoglobin tetramers (HbS) into long filaments
in the deoxy state of the oligomer. The change from a negatively charged
residue to a small hydrophobic one enables a favorable interaction between
hemoglobin subunits from different tetramers. The mutation creates a hydrophobic
patch in subunit b2 (Val6) that
fits to a complementary hydrophobic area between Phe85 and Leu88 in helices
E and F of the b1 subunit that does
not belong to the same tetramer. Fiber growth is initiated by centers
of nucleation and is a function of the concentration of the free HbS tetramer.
Two different nucleation events have been identified, a homogeneous nucleation
from single tetramers, and heterogeneous nucleation on the surface of
preexisting fibers.