Maltoporin
trimer, specific for oligosaccharides (1MAL)
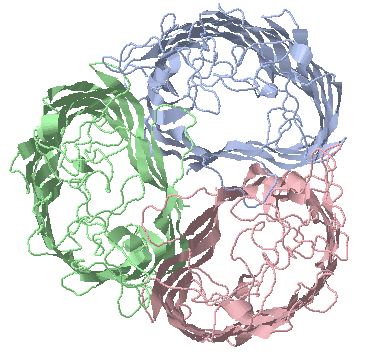
Porins are trimeric outer membrane protein transporters with distinct beta-barrel pore structure in each subunit
a) non-specific trimer (two functional units) OmpF (1OPF)
b) Osmoporin trimer OmpC (2J1N)
c) Phosphoporin monomer PhoE, functional unit is a trimer (1PHO);
d)
trimer, specific for oligosaccharides (1MAL) with bound
sucrose (1AF6) |
Beta-2 Adrenergic Receptor (GPCR) with antibody fragment (24R4); with lysozyme domain (2RH1)
Bacteriorhodopsin, by X-ray diffraction (1AP9) and electron diffraction (1AT9)
Ca-ATPase P-type from muscle sarcoplasmic reticulum membrane (1EUL)
Gap Junction ProteinBeta2 hexamer, connexin 26 hemi-channel or connexon (2ZW)
K-channel
Kcsa bacterial potassium selective channel, closed pore, explained K over Na selectivity (1BL8)
Light
harvesting complex II surround reaction center feeding photon energy to chlorophyll (1KZU)
Reaction
center bacterial photosynthetic reaction center (1PRC)
Rhodopsin, bovine a photosensitive GPCR in the rod cells of the eye at 2.80 Å resolution (1F88)
Channel-forming
Peptides
Gramicidin A
forms head-to-head beta-helix dimer with single pore (1MAG)
Melittin dimer shown; forms helical multimeric barrel (1MLT);
Molecular dynamics
simulation and Channel
recording in artificial membrane
(you need QuickTime3 to view movie)
A complete
list of known structures of membrane proteins at the Stephen
White laboratory at UC Irvine |